1.24.5 Editing Alignment Sequences
- The DNA template sequence in a project with alignments can be edited as usual, however the ClustalW alignment will not be re-run.
Editing the Alignment Sequences
Aligned sequences can only be edited via right click menus in the following ways:
- If an addition exists in the aligned sequence, either select “Add to template” or “Delete from alignment” (Figure 1.24.5.1).
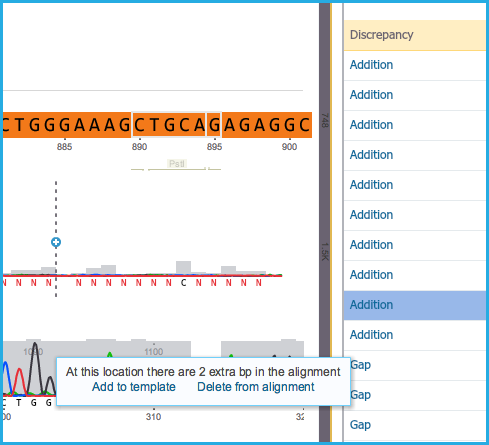 Figure 1.24.5.1: Addition in alignment.
Figure 1.24.5.1: Addition in alignment.</div>
- If a gap exists in the aligned sequence, either select “Add from template” or “Delete from template” (Figure 1.24.5.2).
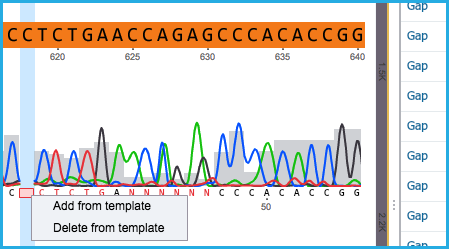 Figure 1.24.5.2: Gap in alignment.
Figure 1.24.5.2: Gap in alignment.</div>
- If a mismatch exists, either select “Copy from template” or “Replace in template” (Figure 1.24.5.3).
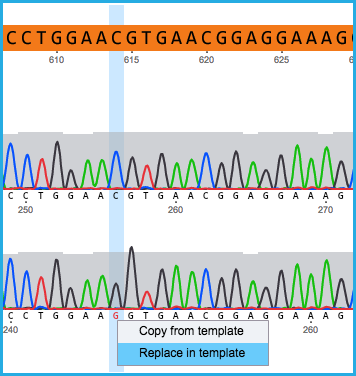 Figure 1.24.5.3: Mismatch in aligment.
Figure 1.24.5.3: Mismatch in aligment.</div>
- If a mismatch exists in terms of an ambiguous base (N) in the alignment, either select “Copy from template” or “Replace to A, T, G or C” (Figure 1.24.5.4).
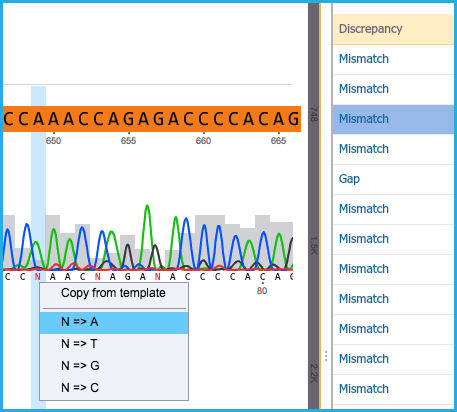 Figure 1.24.5.4: Ambiguous Base.
Figure 1.24.5.4: Ambiguous Base.</div>
Manual Trimming (coming soon)
Sequences can be manually trimmed using the right click drop down menu:
- Trim to.. “Selection” inverts the selection and dulls out the sequence surrounding the selection
- Trim to..”Start” trims and dulls out the sequence up to the left of the caret
- Trim to..”End” trims and dulls out sequence up to the right of the caret
Once the sequence has been trimmed there is a trimming caret which can be dragged back and forth to trim the sequence manually.
If the sequence is on the reverse strand then trimming options work in the reverse.