Transferring DNA/RNA molecule files from SnapGene to Genome compiler
Details about this feature can be found in the main Genome Compiler user guide:
-See section 1.3 for Importing/Exporting files.
First step - Export from Snapgene as a .gb or SnapGene DNA file
Click “File”- “Export sequence” (Figure 3.4.1.1.1) or “Save”/“Save as” (Figure 3.4.1.1.1) in the main menu. Save the file on your computer as a GenBank (.gb/.gbk) file by choosing “GenBank – standard/SnapGene/vector NTI” in the dialogue (Figure 3.4.1.1.1). Alternatively you may save the file as a “SnapGene DNA” file by selecting the appropriate option (Figure 3.4.1.1.2).
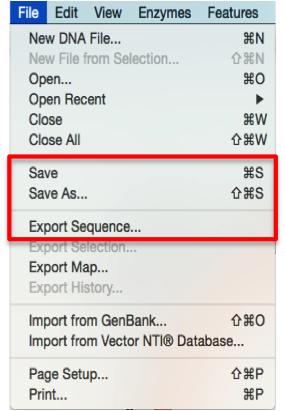 Figure 3.4.1.1.1: Save/Export an open molecule in SnapGene through the main menu.
Figure 3.4.1.1.1: Save/Export an open molecule in SnapGene through the main menu.</div>
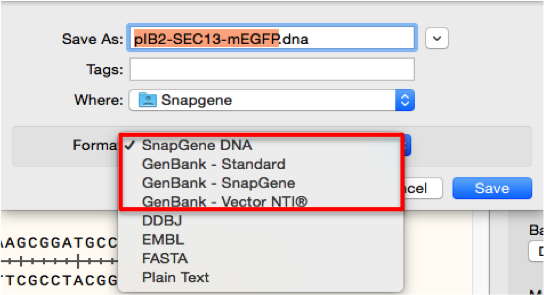 Figure 3.4.1.1.2: Save your molecule as a .gb file at the "Save as" Data window.
Figure 3.4.1.1.2: Save your molecule as a .gb file at the "Save as" Data window.</div>
Second step- Import to Genome Compiler.
To import a file you can either click the “Import File” icon in the main toolbar (Figure 3.4.1.1.3) or in the Materials box (Figure 3.4.1.1.4) or through choosing “File”-“Import” (Figure 3.4.1.1.5). A dialog will pop up to allow you to choose the file from your computer files (Figure 3.4.1.1.6). Once you've selected a file, another dialog will pop up (Figure 3.4.1.1.7) to prompt you for a location. You can choose to save the file in an existing folder or to create a new folder. For more information on importing files see section 1.3.
 Figure 3.4.1.1.3: Import File to Genome Compiler from the icon in the main toolbar
Figure 3.4.1.1.3: Import File to Genome Compiler from the icon in the main toolbar</div>
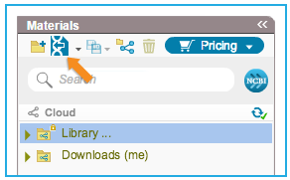 Figure 3.4.1.1.4: Import File to Genome Compiler from the icon in the Materials box
Figure 3.4.1.1.4: Import File to Genome Compiler from the icon in the Materials box</div>
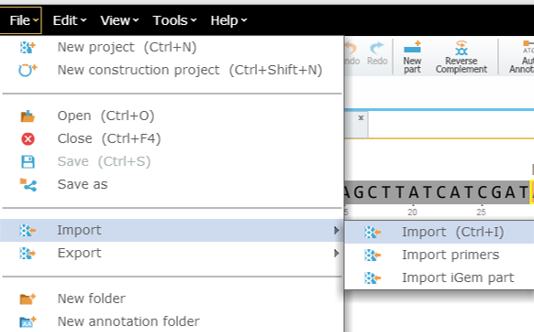 Figure 3.4.1.1.5: Import File to Genome Compiler through choosing “File” in the main menu.
Figure 3.4.1.1.5: Import File to Genome Compiler through choosing “File” in the main menu.</div>
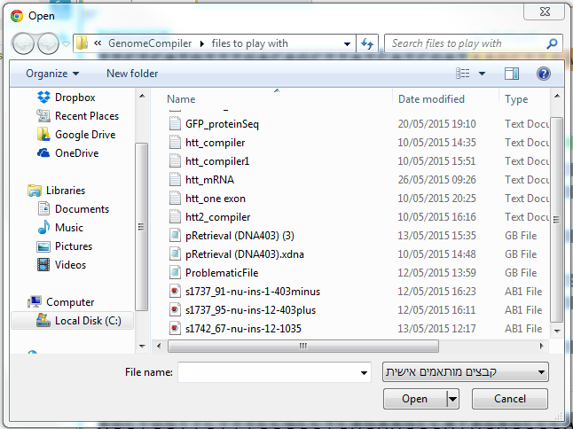 Figure 3.4.1.1.6: Choose the requested file from your computer.
Figure 3.4.1.1.6: Choose the requested file from your computer.</div>
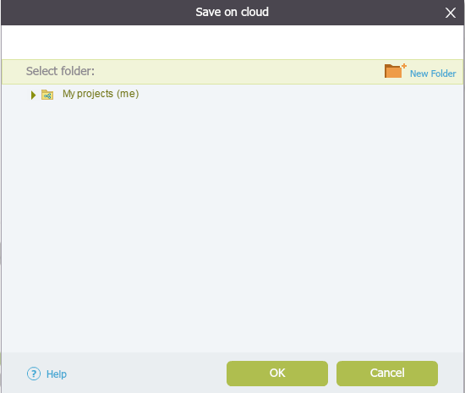 Figure 3.3.1.1.7: Choose the location for the imported file.
Figure 3.3.1.1.7: Choose the location for the imported file.</div>
For more information on Exporting files from Genome Compiler see section 1.3.