1.8.2 Advanced Search
The advanced search dialog allows for more exact controls over the parameters of searching the current project (Figure 1.8.2.1)
 Figure 1.8.2.1: The search bar at the top right of the screen.
Figure 1.8.2.1: The search bar at the top right of the screen.</div>
The dialog is opened by clicking the small gear icon by the search bar (Figure 1.8.2.2) in the top right of the screen and click enter.
 Figure 1.8.2.2: The advanced search dialog is opened by pressing the small gear icon by the quick search bar.
Figure 1.8.2.2: The advanced search dialog is opened by pressing the small gear icon by the quick search bar.</div>
The history of advanced search includes all the history from the quick search bar (Figure 1.8.2.3)
 Figure 1.8.2.3: Advanced search history includes all searches including quick search history.
Figure 1.8.2.3: Advanced search history includes all searches including quick search history.</div>
Define range for search either by selection, specifying exact locations or just searching entire project (Figure 1.8.2.4)
 Figure 1.8.2.4: Search range is defined either by specifying exact base pair location using selection or entire sequence.
Figure 1.8.2.4: Search range is defined either by specifying exact base pair location using selection or entire sequence.</div>
Relax search parameters by defining number of allowed mismatches in sequence (Figure 1.8.2.5)
 Figure 1.8.2.5: Define number of allowed mismatches between sequence and results.
Figure 1.8.2.5: Define number of allowed mismatches between sequence and results.</div>
Decide which alphabet to use while searching, DNA or Amino Acid (Figure 1.8.2.6)
 Figure 1.8.2.6: Define which alphabet to use in search, DNA or Amino Acid.
Figure 1.8.2.6: Define which alphabet to use in search, DNA or Amino Acid.</div>
Define which fields will be searched, including plus and minus strands, project ORFs, primers, annotations, comments, changes, and alignments (Figure 1.8.2.7)
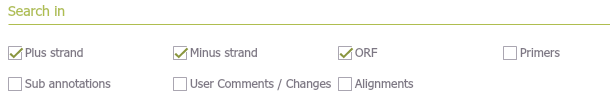 Figure 1.8.2.7: Select which fields to search in the project, strands, ORFs, primers, annotations, comments, changes and alignments.
Figure 1.8.2.7: Select which fields to search in the project, strands, ORFs, primers, annotations, comments, changes and alignments.</div>
All searches will automatically include searching the project and feature properties
Results will appear in the search result tab, as in the quick search.