1.8.1 Searching in Projects
Type a keyword or a DNA sequence in the search bar (Figure 1.8.1.1) in the top right of the screen and click enter.
Figure 1.8.1.1: The search bar at the top right of the screen.</div>
Pressing the arrow on the right will open a list with the history of previous searches (Figure 1.8.1.2)
 Figure 1.8.1.2: Combo box showing the recent search history.
Figure 1.8.1.2: Combo box showing the recent search history.</div>
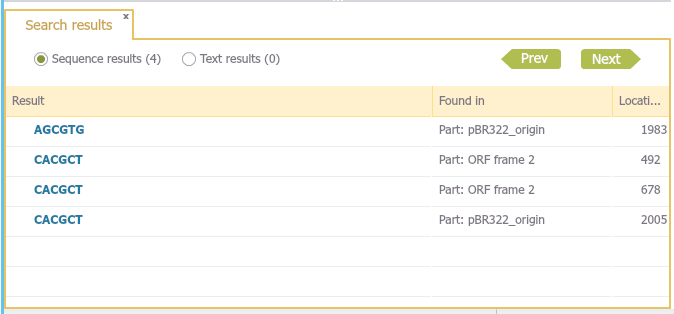
After pressing enter, a tab will open with all search results (Figure 1.8.1.3)
Figure 1.8.1.3: The results tab.</div>
Pressing the "Next" and "Previous" buttons will focus the project on the locations of the results in the all open views.
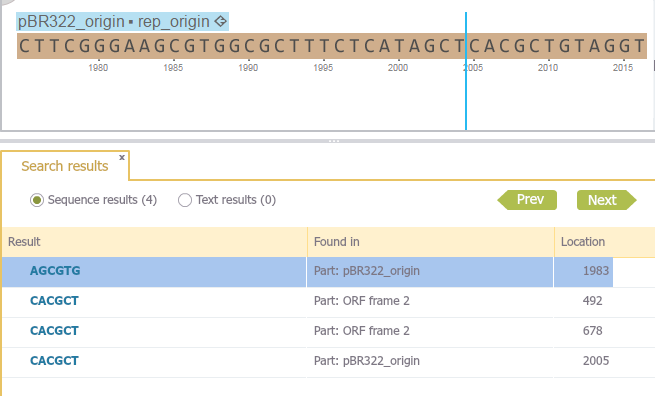
Sequences are searched on both strands using the standard DNA alphabet including ambiguous bases, and Amino Acid alphabet where applicable (Figure 1.8.1.4).
Figure 1.8.1.4: Sequence results in the results tab, sequence query is selected in sequence view.</div>
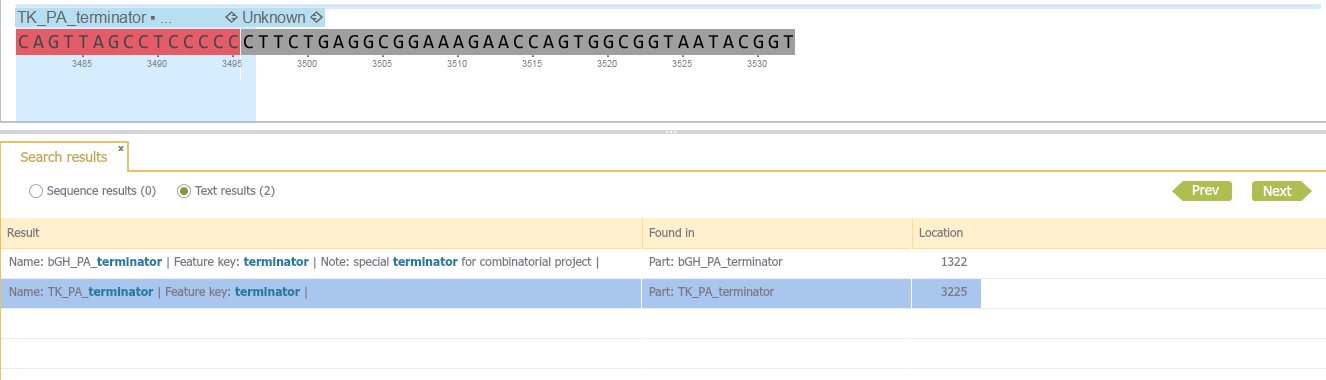
Text searches look for the query in all features metadata, including name, comments, etc. In this case (Figure 1.8.1.5) the word "terminator" appears in two features in multiple fields (Name, Feature Key, Notes)
Figure 1.8.1.5: Text search result in the result tab. The word "terminator" was found in two features in the fields name, feature key, and notes.</div>