3.2.2 Database Explorer a.k.a the Material box
Details about this feature can be found in the main Genome Compiler user guide:
See section 1.4 for importing files.
See section 1.2 for more information on the material box and other user interface properties.
See section 1.7 for Search in the Materials box.
See section 1.9 for search through the NCBI database.
Opening Local Database Explorer:
In Vector NTI you can open the Local database in a new window from the main toolbar (Figure 3.2.2.1) or through “File” in the main menu by choosing “Local Database” (Figure 3.2.2.2).
 Figure 3.2.2.1: Opening Local Database in Vector NTI through the main tool bar.
Figure 3.2.2.1: Opening Local Database in Vector NTI through the main tool bar.</div>
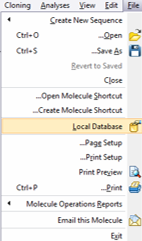 Figure 3.2.2.2: Opening Local Database in Vector NTI through the main tool bar.
Figure 3.2.2.2: Opening Local Database in Vector NTI through the main tool bar.</div>
In Genome Compiler the Databases appear under the “Materials box” to the left of the screen (Figure 3.2.2.3) which can stay open while you work on different projects or be minimized.
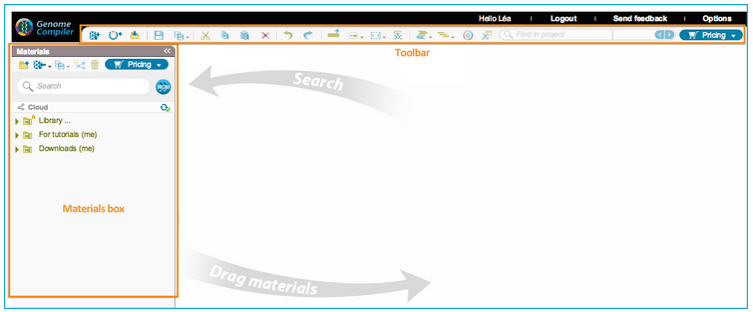 Figure 3.2.2.3: Database displayed in the Material box in Genome Compiler.
Figure 3.2.2.3: Database displayed in the Material box in Genome Compiler.</div>
In Vector NTI the Database Explorer screen consists of several tables which include their predefined libraries (Main) as well as your data in different folders (Figure 3.2.2.4). You can switch between the different tables through the main menu by pressing “Table” and choosing from the drop down menu the appropriate table (Figure 3.2.2.4) or by using the Navigation pane at the bottom left (Figure 3.2.2.5).
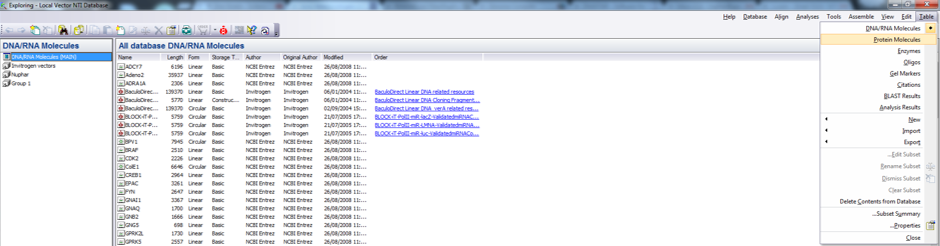 Figure 3.2.2.4: Switching between tables in the local database via the main menu in Vector NTI.
Figure 3.2.2.4: Switching between tables in the local database via the main menu in Vector NTI.</div>
 Figure 3.2.2.5: Navigation pane of the local database in Vector NTI.
Figure 3.2.2.5: Navigation pane of the local database in Vector NTI.</div>
In Genome Compiler all data appears in one place divided to different folders (Figure 3.2.2.6). There are two predefined libraries, one is the “Auto annotation libraries” used for auto annotating your sequences and the second under the name “Library” containing genomes, iGEM & vectors. Upon importing your files (see section 1.4) you can create new folders and sub folders by your request.
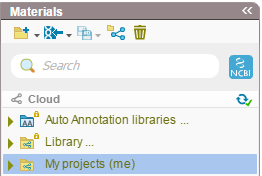 Figure 3.2.2.6: Databases in Genome Compiler.
Figure 3.2.2.6: Databases in Genome Compiler.</div>
For more information regarding the material box and more user interface properties please refer to section 1.2.
In Vector NTI you can reach the search tool through the main tool bar (Figure 3.2.2.7) or through the main menu by going to “Database” and choosing “Search” (Figure 3.2.2.8). The “Molecule Database Search” window will open which looks for your query in the specified folder (Figure 3.2.2.9).
 Figure 3.2.2.7: Opening the Search tool through the main tool bar in Vector NTI.
Figure 3.2.2.7: Opening the Search tool through the main tool bar in Vector NTI.</div>
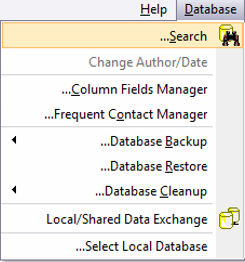 Figure 3.2.2.8: Opening the Search tool through the main menu in Vector NTI.
Figure 3.2.2.8: Opening the Search tool through the main menu in Vector NTI.</div>
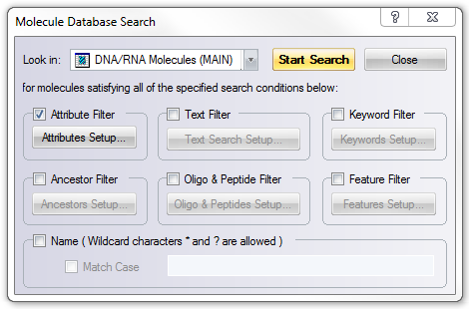 Figure 3.2.2.9: “Molecule Database Search” window in Vector NTI.
Figure 3.2.2.9: “Molecule Database Search” window in Vector NTI.</div>
The search tool in the material box in Genome Compiler (Figure 3.2.2.6) will look for your keyword in the project meta data of all the folders, for more information please refer to section 1.7. Meta data for each project can be found in the “more properties” tab located in a tab in an opened project (Figure 3.2.2.10) or in a tooltip while hovering over the closed project in the Materials box (Figure 3.2.2.11). You can also use it to search through the NCBI database (see section 1.9 for more details) ).
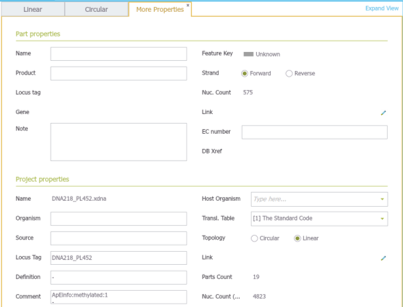 Figure 3.2.2.10: Meta data in the “more properties” tab.
Figure 3.2.2.10: Meta data in the “more properties” tab.</div>
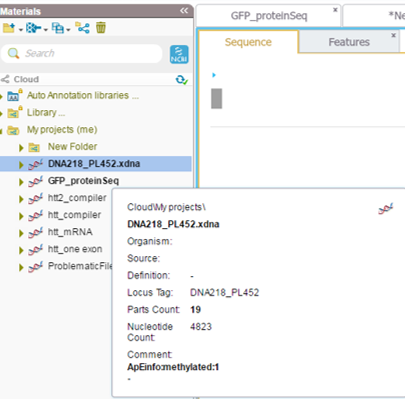 Figure 3.2.2.11: Meta data display of a closed project in the Materials box.
Figure 3.2.2.11: Meta data display of a closed project in the Materials box.</div>