1.25.4 Auto Annotation Settings Dialog
The BLASTN (BLAST "nucleotide") algorithm is used to auto annotate sequences.
Settings Dialogue Overview
Upon selecting to annotate a project, the Auto annotation Settings Dialog (Figure 1.25.4.1) will open where you can:
Select which folders to auto annotate against.
Search according to a stringency; so for example you can run the auto annotation to detect sequences in your project with less than 100% match to those in the library. This will allow you to infer annotations on sequences which are "similar to.." other sequences in the library.
Select "Auto Annotate" and the results will appear in the project.
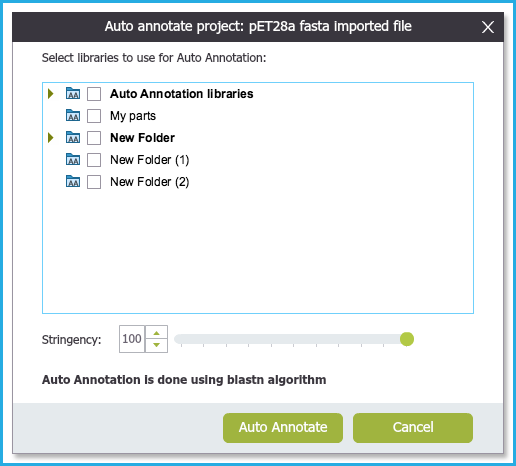 Figure 1.25.4.1: Auto annotation Settings Dialog.
Figure 1.25.4.1: Auto annotation Settings Dialog.</div>
Selecting the appropriate Auto annotation Library
The first step within the settings dialogue is to select which folders/libraries to auto annotate against.
You can select from our internal library taken from "Plasmapper" and/or any other custom libraries which you created.
Use the checkboxes to select or deselect folders.
You can also select sub-folders (Figure 1.25.4.2).
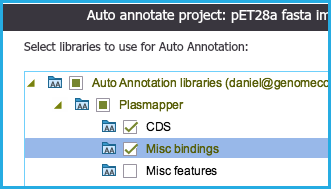 Figure 1.25.4.2: Selecting sub-folders for auto annotation.
Figure 1.25.4.2: Selecting sub-folders for auto annotation.</div>
Selecting the desired Stringency
The next step in the settings dialog is to set the annotation stringency you would like to apply (Figure 1.25.4.3).
Setting a 100% stringency will return annotation results whereby the sequence in the library and the project is exactly the same.
Reducing the stringency will likely return more annotation results with dissimilar sequences between the library and the project but is recommended to infer putative
annotations and functions of the sequence.
 Figure 1.25.4.3: Setting auto annotation stringency.
Figure 1.25.4.3: Setting auto annotation stringency.</div>