Restriction Analysis
Details about this feature can be found in the main Genome Compiler user guide:
-See section 1.16 for Managing Restriction Sites.
-See section 1.17 for Virtual Digest.
In SnapGene you can manage the Restriction Enzymes through the “Enzyme” tab at the bottom of the screen (Figure 3.4.7.1) or from the main menu by opening the “Enzymes” drop down menu (Figure 3.4.7.2). In order to view digestion results you may go to “Tools” from the main menu and select the “Simulate Agarose Gel” to open the digestion simulation window (Figure 3.4.7.4).
 Figure 3.4.7.1: “Enzyme” tab in SnapGene.
Figure 3.4.7.1: “Enzyme” tab in SnapGene.</div>
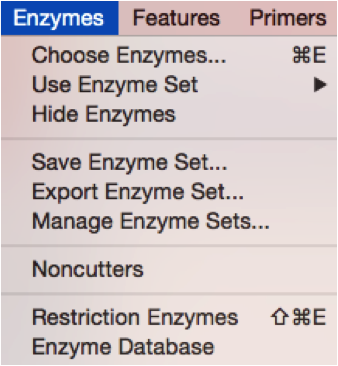 Figure 3.4.7.2: Managing Enzymes from the main menu in SnapGene.
Figure 3.4.7.2: Managing Enzymes from the main menu in SnapGene.</div>
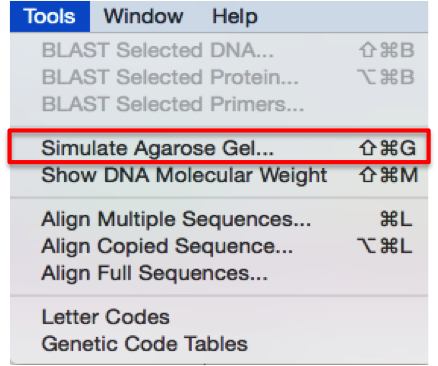 Figure 3.4.7.3: View digestion results in SnapGene.
Figure 3.4.7.3: View digestion results in SnapGene.</div>
In Genome Compiler you can manage restriction enzymes through “Restriction Sites” dialog (Figure 3.4.7.4). You can filter them for different criteria and save presets for further use. To open the “Restriction Sites” dialog, right click on the restriction enzyme in any of the views (Linear/Circular/Sequence) and select the “Restriction settings” from the drop down menu (Figure 3.4.7.5). For more information on managing restriction sites please refer to section 1.16.
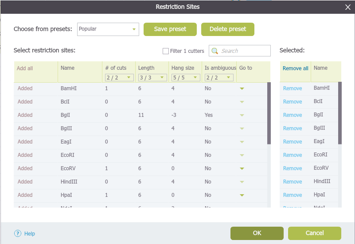 Figure 3.4.7.4: “Restriction Sites” dialog in Genome Compiler.
Figure 3.4.7.4: “Restriction Sites” dialog in Genome Compiler.</div>
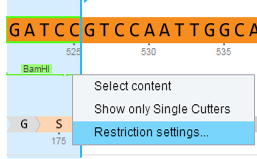 Figure 3.4.7.5: Opening the “Restriction Sites” dialog through right click-“Restriction settings”.
Figure 3.4.7.5: Opening the “Restriction Sites” dialog through right click-“Restriction settings”.</div>
In Genome Compiler in order to perform a hypothetical digest on a sequence selection or on the whole project you should open the “Run Digest” dialog (Figure 3.4.7.7). This dialog can be opened by clicking on “Digest” in the main tool bar (Figure 3.4.7.8) or through opening the “Tools” drop down menu and choosing “Digest” (Figure 3.4.7.9). Alternatively you can right click on project canvas or the project tab and select “Run Digest” (Figure 3.4.7.10). For more information on handling the “Run Digest” dialog please refer to section 1.17.
 Figure 3.4.7.6: “Run Digest” dialog in Genome Compiler.
Figure 3.4.7.6: “Run Digest” dialog in Genome Compiler.</div>
 Figure 3.4.7.7: Opening the “Run Digest” dialog through “Digest” in the main tool bar.
Figure 3.4.7.7: Opening the “Run Digest” dialog through “Digest” in the main tool bar.</div>
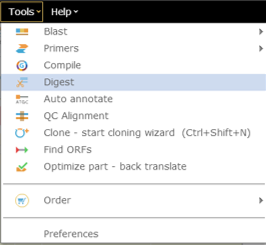 Figure 3.4.7.8: Opening the “Run Digest” dialog through Tools-Digest.
Figure 3.4.7.8: Opening the “Run Digest” dialog through Tools-Digest.</div>
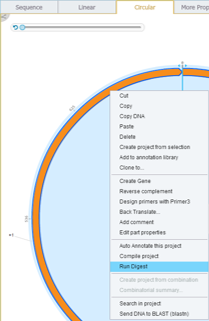 Figure 3.4.7.9: Opening the “Run Digest” dialog through right click-“Run Digest”.
Figure 3.4.7.9: Opening the “Run Digest” dialog through right click-“Run Digest”.</div>