Transferring single/multiple Oligos from SnapGene to Genome compiler
Details about this feature can be found in the main Genome Compiler user guide:
-See section 1.3 for Importing/Exporting files.
-See section 1.18.5 for Primer Summary Table information.
-See section 1.18 for Primer Design.
Export from SnapGene as csv or txt file..
In SnapGene first select the required primer and then open the “Primers” drop down menu from the main menu, choose “Export Selected Primer...” (Figure 3.4.1.3.1) and then “Comma-separated values” or “Tab- delimited values” for the file format (Figure 3.4.1.3.2). Both formats creates a file with two columns, the first one is the primer’s name and the second is it’s sequence. These files can be directly imported to Genome Compiler as explained below.
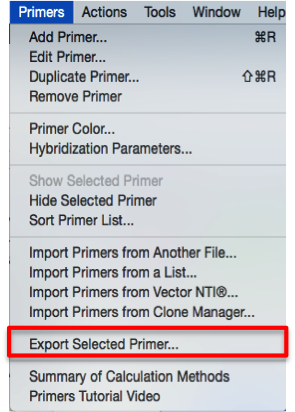 Figure 3.4.1.3.1: Export primers using the main menu in SnapGene.
Figure 3.4.1.3.1: Export primers using the main menu in SnapGene.</div>
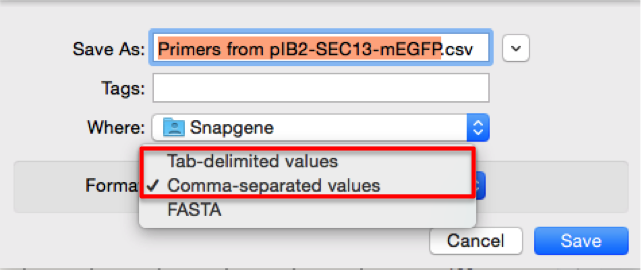 Figure 3.4.1.3.2: Choose “Comma-separated values” or “Tab- delimited values” when exporting primers from SnapGene.
Figure 3.4.1.3.2: Choose “Comma-separated values” or “Tab- delimited values” when exporting primers from SnapGene.</div>
If you already have a csv/txt primers file you can import many primers at once, just be sure to rearrange the list in the following columns order (Figure 3.4.1.3.3):
NAME/SEQUENCE/STRAND/LEADER (e.g Primer1,TGACGATGCCGATTTGGAAA,PLUS,GCGCG)
Comma separated: NAME,SEQUENCE,STRAND,LEADER (e.g Primer1,TGACGATGCCGATTTGGAAA,MINUS,GCGCG)
Tab separated:NAME SEQUENCE STRAND LEADER (e.g Primer1 TGACGATGCCGATTTGGAAA PLUS GCGCG)
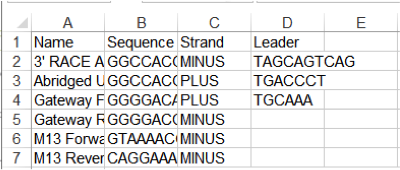 Figure 3.4.1.3.3: Primers table example in Excel.
Figure 3.4.1.3.3: Primers table example in Excel.</div>
Import primers table to Genome Compiler.
Starting with your target project open, navigate to the menu bar and select “File,” then “Import,” and then “Import Primers” (Figure 3.4.1.3.4). The “Import Primer” dialog box will then open where you can upload your primer library (Figure 3.4.1.3.5). Select “choose a file” to choose the appropriate file. The primers will then be imported into the Import Primers dialog where you can select the checkboxes of the primers you would like to detect in the project and then “Import”. The primers which are complementary to your sequence will appear on the sequence (Figure 3.4.1.3.6) as well as in the Primer Summary Table for that specific project (Figure 3.4.1.3.7) (see section 1.18.5. for Primer Summary Table information).
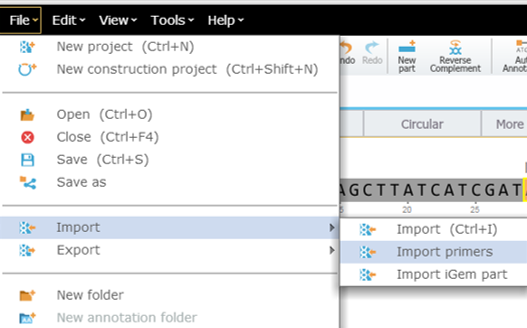 Figure 3.4.1.3.4: Opening "Import Primers" dialog box in Genome Compiler.
Figure 3.4.1.3.4: Opening "Import Primers" dialog box in Genome Compiler.</div>
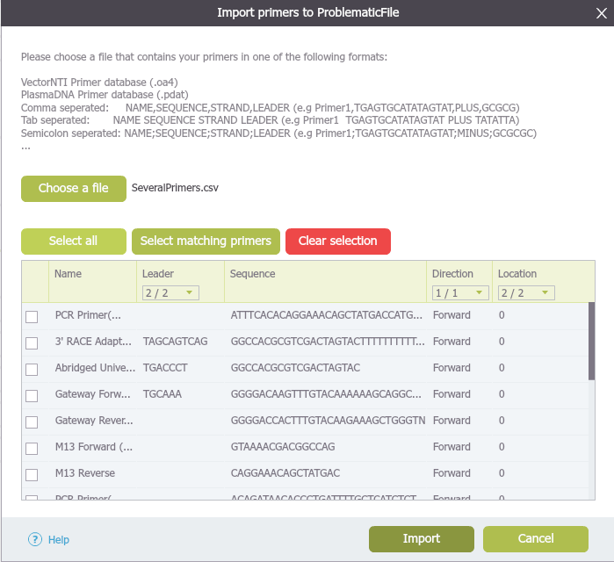 Figure 3.4.1.3.5: Upload your primer library through the "Import Primers" dialog box.
Figure 3.4.1.3.5: Upload your primer library through the "Import Primers" dialog box.</div>
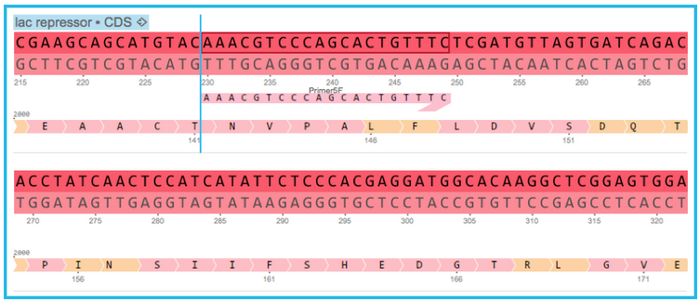 Figure 3.4.1.3.6: Complementary primers will appear in the sequence view.
Figure 3.4.1.3.6: Complementary primers will appear in the sequence view.</div>
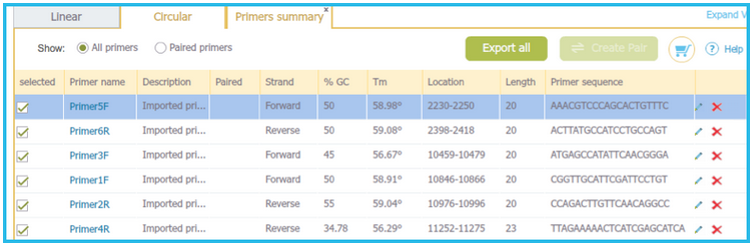 Figure 3.4.1.3.7: Complementary primers will appear in the project Primer Summary Table.
Figure 3.4.1.3.7: Complementary primers will appear in the project Primer Summary Table.</div>
Note: If the primers you imported are not complementary to anywhere on the sequence then they will not be added to your project or saved anywhere inside the program. For more information on importing/exporting files please refer to section 1.3.