Transferring Protein molecule files from SnapGene to Genome compiler
Details about this feature can be found in the main Genome Compiler user guide:
-See section 1.3 for Importing/Exporting files.
First step- Export from SnapGene as .FASTA file.
Export from Snapgene by clicking “File”- “Export sequence” (Figure 3.4.1.2.1) or “Save”/“Save as” (Figure 3.4.1.2.1) through the main menu. Save the file on your computer as a FASTA (.fa) file by choosing “FASTA” in the dialogue (Figure 3.4.1.2.1).
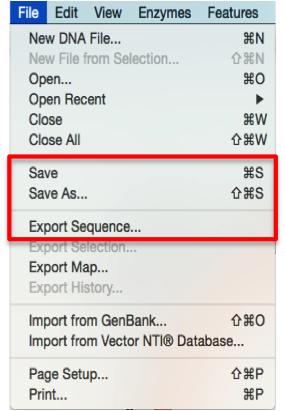 Figure 3.4.1.2.1: Save/Export an open molecule in SnapGene through the main menu.
Figure 3.4.1.2.1: Save/Export an open molecule in SnapGene through the main menu.</div>
 Figure 3.4.1.2.2: Save your molecule as a .FASTA file at the "Save as" Data window.
Figure 3.4.1.2.2: Save your molecule as a .FASTA file at the "Save as" Data window.</div>
Second step- Import to Genome Compiler.
To import a file you can either click the “Import File” icon in the main toolbar (Figure 3.4.1.2.3) or in the Materials box (Figure 3.4.1.2.4) or through choosing “File”-“Import” (Figure 3.4.1.2.5). A dialog will pop up to allow you to choose the file from your computer files (Figure 3.4.1.2.6). Once you've selected a file, another dialog will pop up (Figure 3.4.1.2.7) to prompt you for a location. You can choose to save the file in an existing folder or to create a new folder.
For more information on importing files see section 1.3.
 Figure 3.4.1.2.3: Import File to Genome Compiler from the icon in the main toolbar
Figure 3.4.1.2.3: Import File to Genome Compiler from the icon in the main toolbar</div>
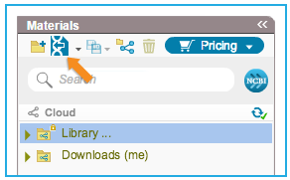 Figure 3.4.1.2.4: Import File to Genome Compiler from the icon in the Materials box
Figure 3.4.1.2.4: Import File to Genome Compiler from the icon in the Materials box</div>
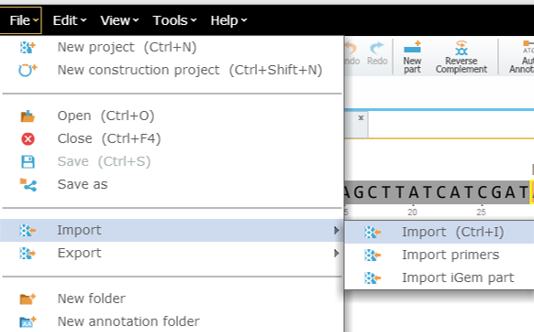 Figure 3.4.1.2.5: Import File to Genome Compiler through choosing “File”-“Import”
Figure 3.4.1.2.5: Import File to Genome Compiler through choosing “File”-“Import”</div>
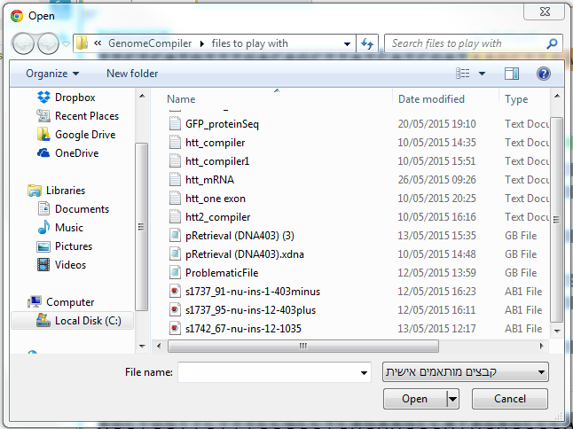 Figure 3.4.1.2.6: Choose the requested file from your computer
Figure 3.4.1.2.6: Choose the requested file from your computer</div>
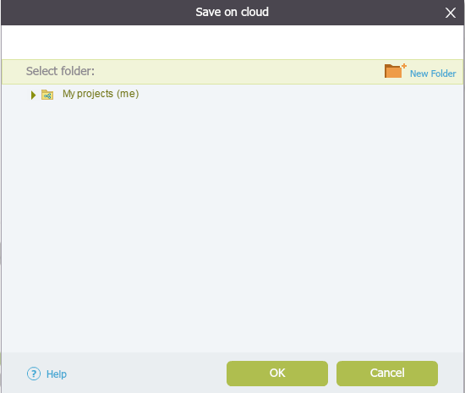 Figure 3.4.1.2.7: Choose the location for the imported file
Figure 3.4.1.2.7: Choose the location for the imported file</div>
For more information on Exporting files from Genome Compiler see section 1.3.
In case you saved your molecule as a DNA sequence, you may upload it as a DNA file (as explained in section 3.4.1.1) and translate your DNA sequence to protein.
Choose the whole sequence or a fragment and set as CDS by first selecting the sequence in one of the Sequence/Circular/Linear views. Then you can right click and either choose “Annotate selection” (Figure 3.4.1.2.8) or “Edit part properties” (Figure 3.4.1.2.9) or directly go to the “More Properties” tab (Figure 3.4.1.2.10). In this tab open the drop down “Feature Key” window and choose CDS (or all other orange feature keys) (Figure 3.4.1.2.10). This will automatically translate your sequence and display it underneath the DNA sequence (Figure 3.4.1.2.11).
 Figure 3.4.1.2.8: Setting selection as CDS through right click-“Annotate selection”.
Figure 3.4.1.2.8: Setting selection as CDS through right click-“Annotate selection”.</div>
 Figure 3.4.1.2.9: Setting selection as CDS through right click-“Edit part properties”.
Figure 3.4.1.2.9: Setting selection as CDS through right click-“Edit part properties”.</div>
 Figure 3.4.1.2.10: “More Properties” tab.
Figure 3.4.1.2.10: “More Properties” tab.</div>
 Figure 3.4.1.2.11: Protein sequence display underneath the DNA sequence.
Figure 3.4.1.2.11: Protein sequence display underneath the DNA sequence.</div>