Find a.k.a Search in Project
Details about this feature can be found in the main Genome Compiler user guide:
-See section 1.7 for search in project.
In Snapgene to search a sequence or an Enzyme/Feature/Primer in the project you should either press ctrl+f or select “Edit”- “Find” from the main menu (Figure 3.4.4.1). This will open the “Find” panel in which you may specify your search term (Figure 3.4.4.2).
 Figure 3.4.4.1: Opening “Find” panel in SnapGene through the main menu.
Figure 3.4.4.1: Opening “Find” panel in SnapGene through the main menu.</div>
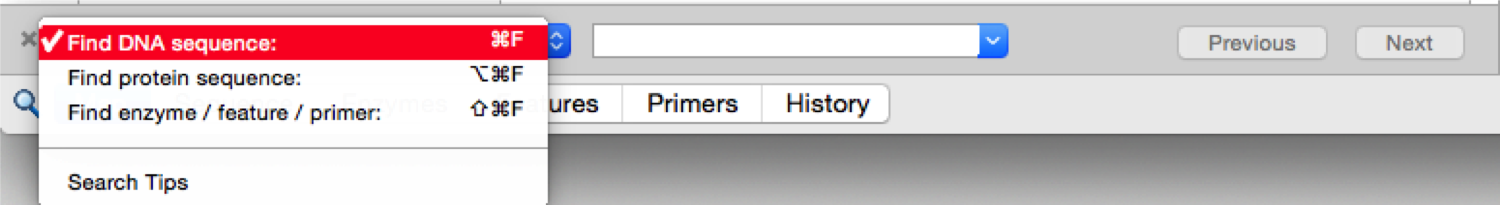 Figure 3.4.4.2: “Find” panel in SnapGene.
Figure 3.4.4.2: “Find” panel in SnapGene.</div>
In Genome Compiler you may type a DNA sequence or a keyword in the search bar (Figure 3.4.4.3) in the top right of the screen and click enter, the search results will be displayed in a new tab. Pressing the arrow on the right will open a list with the history of previous searches (Figure 3.4.4.4). There is no need to select a strand since Genome Compiler looks for the sequence in both strands using the standard DNA alphabet including ambiguous bases, and Amino Acid alphabet where applicable. When you search text, the software looks for the query in all features metadata. The advanced search dialog (Figure 3.4.4.5) allows for more exact controls over the parameters of searching the current project. To open the advanced search dialog you can either press ctrl+f or click the small gear icon at the top right of the search bar (Figure 3.4.4.6). For more information regarding search in project, please refer to section 1.7.
Figure 3.4.4.3: The search bar in Genome Compiler.</div>
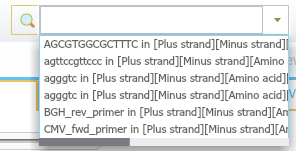 Figure 3.4.4.4: Opening history of previous searches.
Figure 3.4.4.4: Opening history of previous searches.</div>
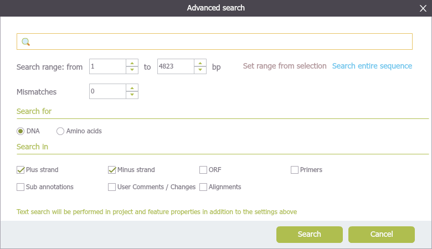 Figure 3.4.4.5: Advanced search dialog.
Figure 3.4.4.5: Advanced search dialog.</div>