3.3.8 ORF Detection
Details about this feature can be found in the main Genome Compiler user guide:
See section 1.23 for ORF Detection.
To identify ORFs in ApE you should go to “ORF” in the main menu and choose your parameters in the drop down menu (Figure 3.3.8.1). To see the results you can either click “Find next/previous” to display one ORF at a time on the sequence or choose “ORF Map” (Figure 3.3.8.1) to view all ORFs (Figure 3.3.8.2).
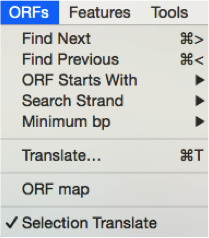 Figure 3.3.8.1: ORF tools in ApE from the main menu.
Figure 3.3.8.1: ORF tools in ApE from the main menu.</div>
 Figure 3.3.8.2: ORF Map in ApE.
Figure 3.3.8.2: ORF Map in ApE.</div>
In Genome Compiler Before you run the ORF detection, you may wish to adjust the detection settings (Figure 3.3.8.3) (See section 1.23). You can access the ORF settings dialog in two ways: Either from the menu bar, by selecting "View“-"Settings“-"ORF“ (Figure 3.3.8.4) or from the Annotation Layers menu at the bottom right panel of the opened project and selecting the ORF settings icon (Figure 3.3.8.5).After adjusting the ORF detection settings click OK to start the detection. The ORFs will appear in the linear/circular views as arrows and in the sequence view as a AA sequence beneath the appropriate DNA fragment (Figure 3.3.8.6).
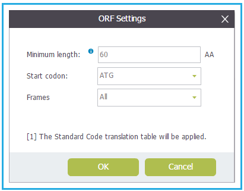 Figure 3.3.8.3: ORF detection settings dialog in Genome Compiler.
Figure 3.3.8.3: ORF detection settings dialog in Genome Compiler.</div>
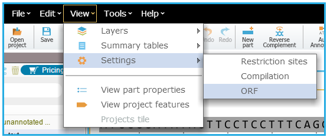 Figure 3.3.8.4: Accessing the ORF settings dialog from the menu bar in Genome Compiler.
Figure 3.3.8.4: Accessing the ORF settings dialog from the menu bar in Genome Compiler.</div>
 Figure 3.3.8.5: Accessing the ORF settings dialog from the Annotation Layers menu in Genome Compiler.
Figure 3.3.8.5: Accessing the ORF settings dialog from the Annotation Layers menu in Genome Compiler.</div>
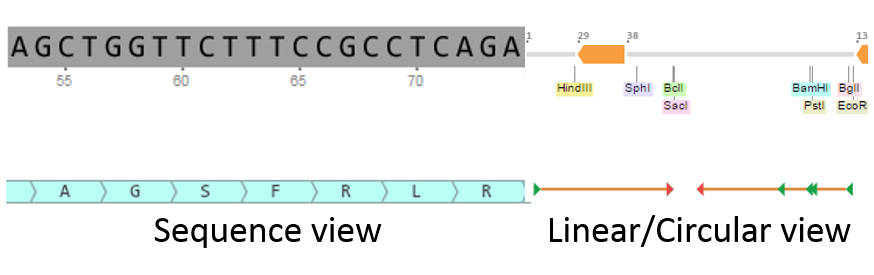 Figure 3.3.8.6: New ORFs in Genome Compiler.
Figure 3.3.8.6: New ORFs in Genome Compiler.</div>